Rival Scientists Cast Doubt Upon Recent Discovery About Invincible Animals
A recent claim that tardigrades got a sixth of their DNA from microbes is starting to unravel.
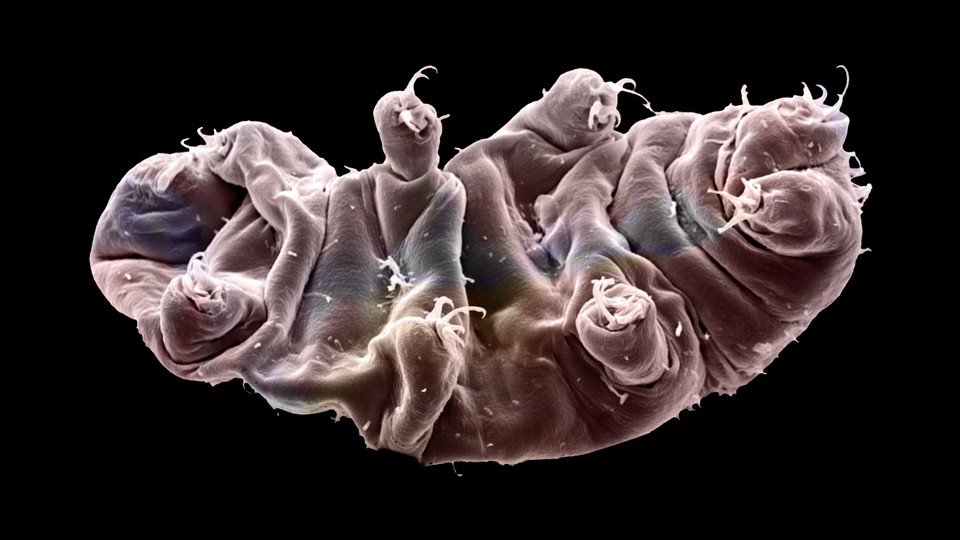
Last Monday, a team from the University of North Carolina at Chapel Hill published the first ever genome of a tardigrade—a group of endearing microscopic animals with a reputation for being nigh-invincible. Astonishingly, as we reported last week, they found that around 6,600 of the animal’s genes—a full sixth of its genome—had jumped in from bacteria and other foreign sources. And perhaps, they speculated, this massive horizontal gene transfer (HGT) explained the tardigrade’s famed ability to withstand extreme conditions.
Just one week later, those claims are starting to unravel. A second team from the University of Edinburgh had also been sequencing the genome of the same species of tardigrade, ordered from the same supplier. And their results, released on Tuesday as a preprint paper, are totally different.
They found very few horizontally transferred genes—as few as 36, and just 500 at the very most. They concluded that their rivals had sequenced DNA from bacteria that were living alongside the tardigrades and, despite their best efforts, had mistaken the genes of those microbes for genuine tardigrade genes.
These disputes arise because, until recently, scientists didn’t have the technology to sequence genomes continuously, so they had to break the DNA into small pieces. They could then sequence these fragments individually and assemble the small “reads” into a coherent whole.
When you do this, you ought to get more or less the same number of reads for every gene in an animal’s genome. But when the Edinburgh team looked at their own data, they saw that many reads were incredibly rare while others were up to 10 times as common. “There is no way, biologically, these can be part of the same genome,” says Mark Blaxter, who led the team. Instead, the rare reads probably came from stowaway bacteria. The team carefully cleaned up their data to remove these contaminating sequences.
They ended up with around 500 genes that potentially came from microbes, and they still think that the most of these are from contaminants that they haven’t been able to sort through yet. They only have strong evidence for 36 genes being horizontally transferred from bacteria, which is within the common range for animal genomes.
The Edinburgh team were still polishing their data when the rival paper came out last Monday, claiming 6,600 horizontally transferred genes. They were shocked, but rushed to analyze the data that Bob Goldstein from UNC quickly uploaded onto a server.
Worryingly, they found that the UNC data included many reads that they hadn’t seen at all—even though both groups sequenced the same animals! And most of these phantom reads were rare. Based on this, the Edinburgh team concluded that around 30 percent of the UNC genome probably came from contaminating microbes.
“If this is true, it is damning,” says John McCutcheon from the University of Montana. “But it’s also surprising, because much of what [the UNC team] did was pretty careful, so I would not have expected them to miss this.”
“We thought seriously about the possibility of contamination—it was of course the most likely initial explanation for the large amount of foreign DNA found in our assembly—and much of the analysis in our paper was designed specifically to address this issue,” says Thomas Boothby from the UNC team, in a comment.
For example, the UNC team focused on 107 parts of their assembled genome where a gene of bacterial origin seemed to sit next to one of animal origin. They used a technique that started with DNA sequences at either end of these pairs, and tried to duplicate all the DNA in between. If the technique worked—and it largely did—it would mean that both genes really are connected on the same stretch of DNA, and that the bacterial one couldn’t have come from contaminants. What’s more, 54 of these 107 genes were among the 500 that the Edinburgh team had singled out as being potential horizontal transfers.
Much depends on whether these 107 pairs are representative of the 6600 genes that were supposedly transferred from bacteria. The UNC are treating them as such. The Edinburgh team thinks they can’t possibly be. David Baltrus from the University of Arizona, who wasn’t involved with either group, says, “My gut is telling me that the UNC authors somehow, probably unintentionally, biased picking this random subset.”
The UNC team also used a different sequencing system called PacBio, which decodes strands of DNA without first breaking them. This supposedly revealed that the foreign genes are physically linked to the tardigrade’s native ones. These claims were hard for the Edinburgh team to assess, since the UNC group hadn’t released their PacBio data at the time; they have since done so.*
“I want to believe that massive HGT happened, because it would be an awesome story,” says Baltrus. “But the problem is that extraordinary claims require extraordinary evidence.”
And there are other reasons to doubt the UNC conclusion. They estimated that the tardigrade’s genome contained 250 million DNA “letters.” The Edinburgh group thought the same initially, but after removing the potential bacterial contaminants, they got a much slimmer genome with just 135 million letters. That’s much more in line with predictions from other scientists, based on the size of the animal’s cells.
This isn’t the first time that claims of horizontal gene transfer have unravelled. When the first draft of the human genome was published in 2001, the scientists behind the project said that they had identified 223 genes that “appear likely to have resulted from horizontal transfer from bacteria”—a claim that evaporated after more critical analysis. All of this shows just how difficult it can be to decipher and interpret genomes, even in an age when it seems commonplace to do so.
McCutcheon says that it would be useful to sequence the genomes of other tardigrades. If the alleged horizontally transferred genes exist in other species, “it would be pretty compelling evidence that they are real.”
Meanwhile, Sujai Kumar from the Edinburgh team says, “The entire process is a victory for open science.” His colleagues could never have done their analysis if their rivals hadn’t willingly and promptly released their data. And even just a few years ago, they would have had nowhere to upload a manuscript detailing the conflicting results, which other scientists could check and discuss. It took just nine days for the second paper to follow the first.
“What is evident is the amazing new ability of science to self correct rapidly,” says Blaxter. “What would in decades past have taken many months to sort out was immediately the focus of bright minds across the planet, asking questions, requesting data, speculating on the possible new biology, and collectively making sure that our science deals in validated inference.”
* This article originally stated that the UNC group had not released its PacBio data yet. We regret the error.